Quaternary Structure Collection
"Unveiling the Intricacies of Quaternary Structure: From Anaesthetic Inhibition to Tumour Suppression" The quaternary structure
All Professionally Made to Order for Quick Shipping
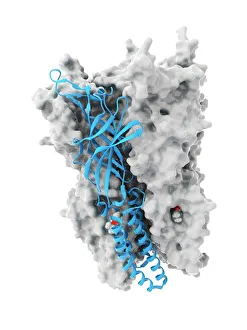
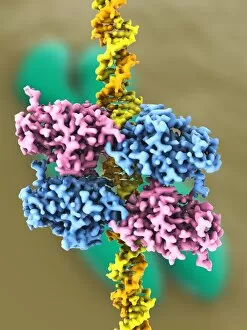
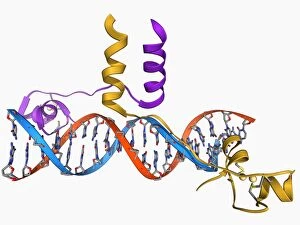
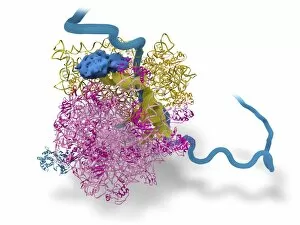
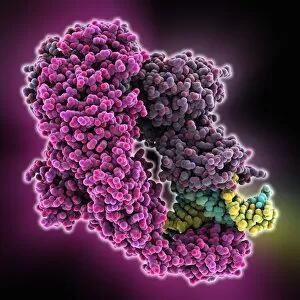
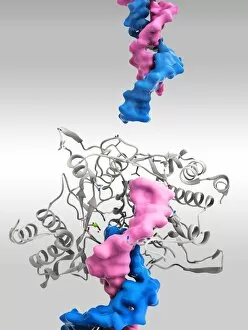
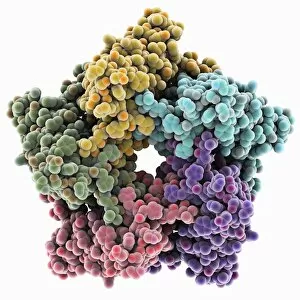
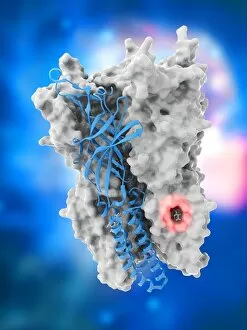
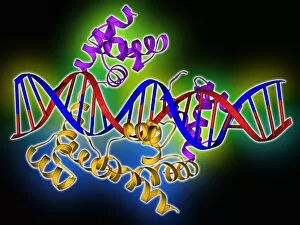
"Unveiling the Intricacies of Quaternary Structure: From Anaesthetic Inhibition to Tumour Suppression" The quaternary structure, a fascinating aspect of protein organization, plays a crucial role in various biological processes. One such example is seen with an anaesthetic inhibiting an ion channel (C015/6718), where the intricate arrangement of subunits determines its functionality. In the realm of cancer research, we delve into the interaction between tumour suppressor proteins and DNA (C017/3647). Understanding their quaternary structure sheds light on how these proteins regulate cell growth and prevent tumor formation. Similarly, exploring this relationship uncovers potential therapeutic targets for combating cancer. Artwork depicting Ricin A-chain (C017/3653) and Ricin molecule (C017/3652) showcases another intriguing facet of quaternary structure. These illustrations highlight how multiple subunits come together to form a deadly toxin or enzyme that can be harnessed for medical advancements. Continuing our exploration into tumour suppression mechanisms, we encounter further instances where tumour suppressor proteins interact with DNA (C017/3644 & C017/3646). The intricacies within their quaternary structures provide insights into genetic regulation and offer potential avenues for targeted therapies against malignancies. Moving beyond cancer-related studies, we encounter HK97 bacteriophage procapsid formations - examples that showcase viral capsids' complex assembly process. Investigating their quaternary structures allows us to understand viral infection mechanisms better while potentially identifying novel antiviral strategies. Rounding out our journey through diverse fields is artwork portraying Ricin A-chain (C017/3654), which emphasizes the importance of understanding protein architecture in designing countermeasures against biothreats like bioterrorism agents.